|
Vertical Divider
|
Genomics of tolerance to avian malaria among Hawaii amakihi: Avian malaria was introduced to Hawaii in the 1930s and caused devastating population declines in many native Hawaiian birds, driving some to extinction. Some species, notably the Hawai’i 'amakihi (Chlorodrepanis virens), were able to persist in small, isolated populations and have developed the ability to survive malaria infection. Recently, amakihi populations have begun to rebound despite the widespread presence of malaria. It is our goal to characterize the genomic changes that are involved in resistance and/or tolerance to avian malaria.
|
|
Low-elevation populations have evolved under the recent selection pressure of avian malaria, whereas high-elevation populations (which are too cold for the mosquito vector) have not. Because amakihi individuals disperse much farther within an elevation than across elevation, there is genetic differentiation between low- and high-elevation populations (right, differentiation between one low- and one high-elevation population). |
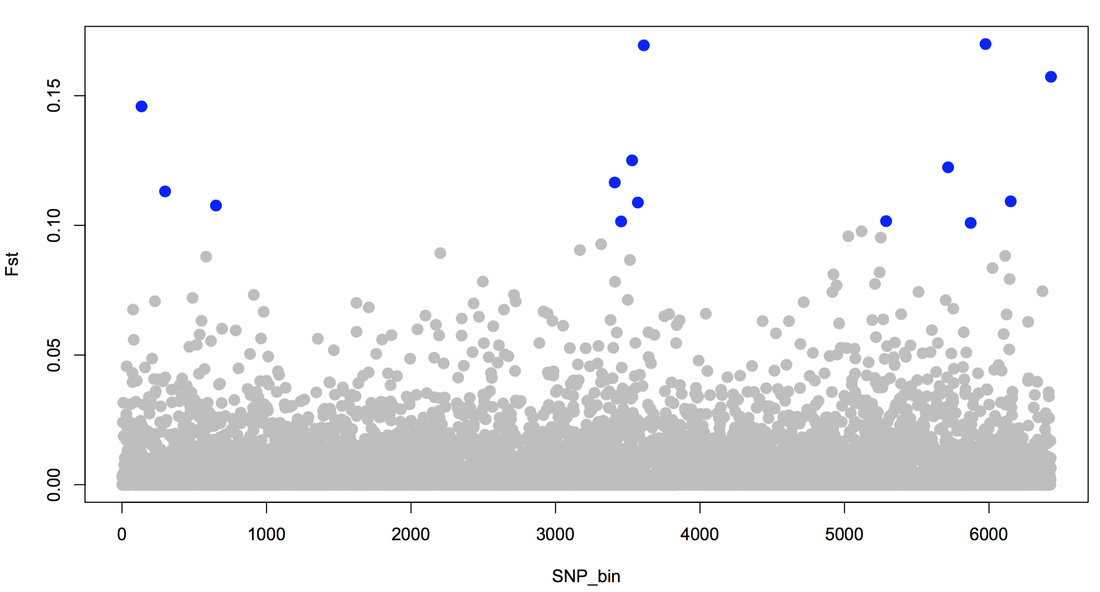
Along with other approaches, we use Fst-outlier tests to detect loci that are differentiated between elevations more than the background rate of differentiation. We combine this with direct tests of selection (e.g., Tajima’s D and selective sweeps) to identify loci in common to all methods. We complement this spatial approach with genome-wide association methods to detect sites that are associated with survivorship from malaria in experimental infection studies, and RNA-seq on both experimentally infected and spatially sampled birds.
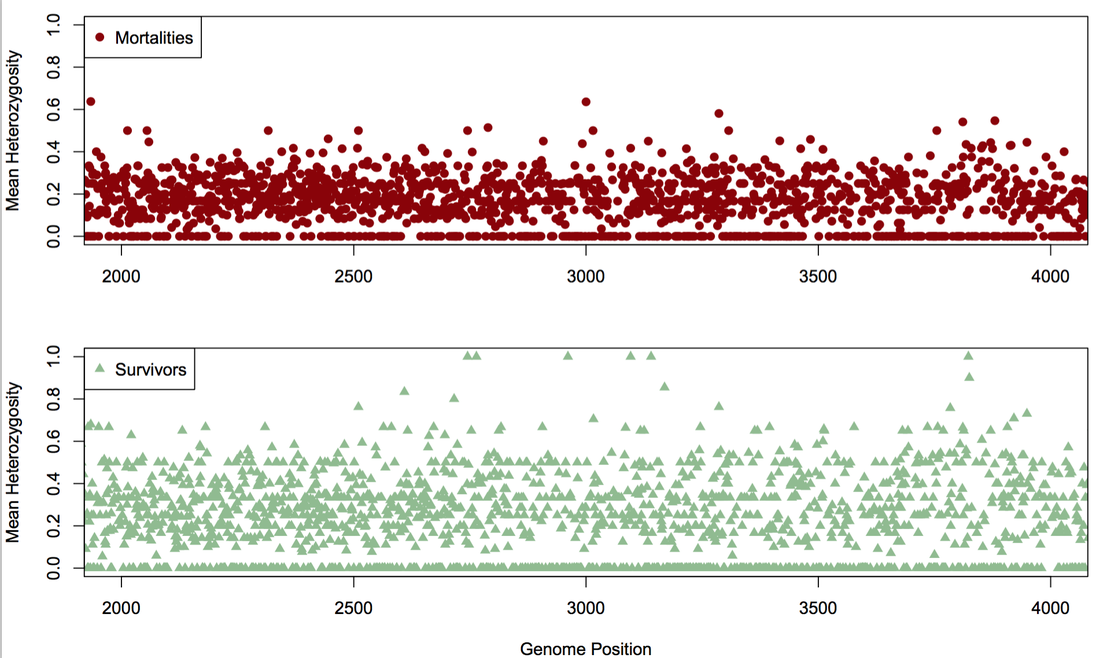
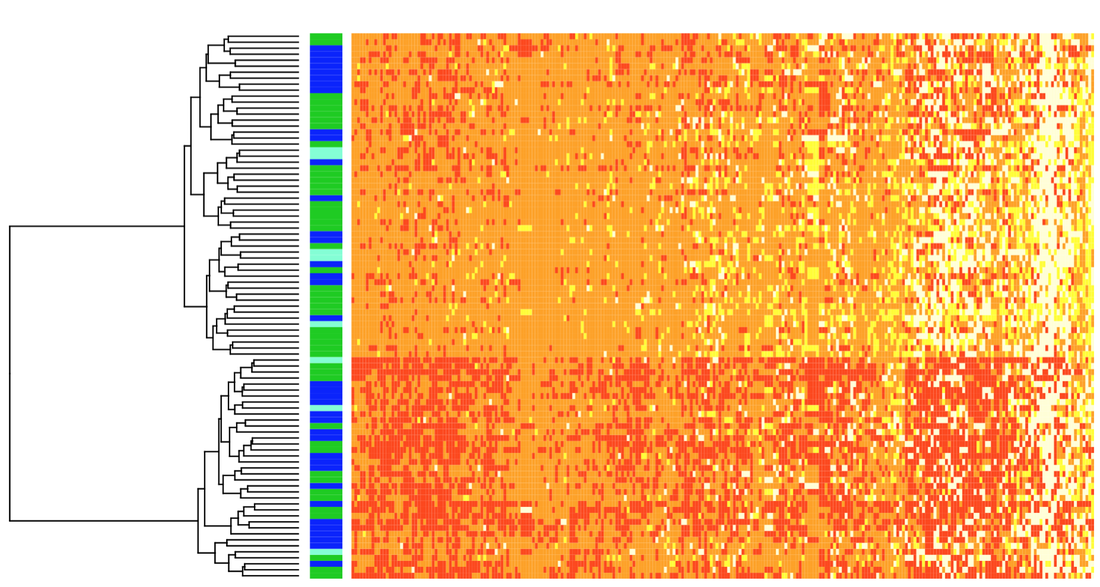
Below (left), heterozygosity of experimentally-infected mortalities (top panel) and survivors (bottom panel) is depicted in one section of the genome. Survivors had higher heterozygosity than mortalities, and this pattern was consistent across the genome. Below right: 259 SNPs (columns) were fixed for different alleles (each warm color) in mortalities and survivors from the experimental challenge. We genotyped individuals in natural populations at these SNPs and saw clustering between low-elevation (green horizontal bars) and high-elevation (dark blue horizontal bars; light blue are mid-elevation) populations.
Below (left), heterozygosity of experimentally-infected mortalities (top panel) and survivors (bottom panel) is depicted in one section of the genome. Survivors had higher heterozygosity than mortalities, and this pattern was consistent across the genome. Below right: 259 SNPs (columns) were fixed for different alleles (each warm color) in mortalities and survivors from the experimental challenge. We genotyped individuals in natural populations at these SNPs and saw clustering between low-elevation (green horizontal bars) and high-elevation (dark blue horizontal bars; light blue are mid-elevation) populations.
Vertical Divider
Good news: We just found out that our EEID proposal was funded to extend this work to understand a more holistic picture of the evolution of avian hosts, mosquito vectors, and the parasite on the Hawaiian Islands. The project includes spatial and temporal sampling of the three types of taxa and an integrative model tying the pieces together into a cohesive picture of evolution in a disease system. Collaborators include Dina Fonseca, Nina Fefferman, Rob Fleischer, Jeff Foster, Marm Kilpatrick, and Eben Paxton.
Collaborators:
Carter Atkinson, USGS Pacific Islands Research Institute
Rob Fleischer, Smithsonian National Zoo
Eben Paxton, USGS Pacific Islands Research Institute
Kristina Paxton, University of Hawaii-Hilo